What Can Be Learned from Cryogenic Electron Microscopy and Tomography from Biological Samples?
Prof. Wah Chiu, Stanford University
Abstract
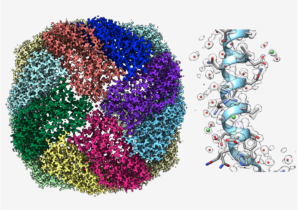
Cryogenic electron microscopy (cryoEM) was introduced almost 50 years ago in an attempt to preserve structural integrity of biological samples to keep them in a frozen, hydrated state in an electron microscope vacuum [1] and to reduce electron radiation damage in a transmission electron microscope [2]. The freezing protocol was subsequently improved by plunge freezing so that the frozen sample can be vitrified and fully embedded in a thin layer of vitreous ice [3]. In the ensuing years, numerous investigations have been conducted to improve the quality of image data of vitrified biological samples by optimizing the cryo-specimen stage temperature choice [4], using direct electron counting device for image recording [5] and reducing beam-induced specimen movement effects by computationally aligning multiple images from the same specimen area [6]. Since an electron image is a 2-dimensional projection of a 3-dimensional electrostatic potential, reconstruction from multiple views of the samples allows a visualization of the object in 3-dimension [7]. Years of implementation of software from various groups [8, 9, 10] have provided the end-users open-source software to determine near to true atomic resolution 3-D structures of single macromolecular complexes even with compositional and conformational heterogeneity [11]. Another emerging technology is to use cryogenic electron tomography to study cells in cellulo [12]. This later approach requires the use of a focused ion beam to mill vitrified cells with tens of microns in thickness to thin lamellae so that tomographic series can be recorded and reconstructed [13]. All these advances in biological cryogenic electron microscopy have inspired its applications to characterize structures of materials used for energy production [14, 15]. We can expect that this cryogenic electron imaging approach will make many new discoveries in energy materials in the coming years.
1. Taylor KA, Glaeser RM. Electron Diffraction of Frozen, Hydrated Protein Crystals. Science. 1974. pp. 1036–1037. doi:10.1126/science.186.4168.1036 2. Hayward SB, Glaeser RM. Radiation damage of purple membrane at low temperature. Ultramicroscopy. 1979. pp. 201–210. doi:10.1016/s0304-3991(79)90211-0 3. Dubochet J, Lepault J, Freeman R, Berriman JA, Homo J-C. Electron microscopy of frozen water and aqueous solutions. Journal of Microscopy. 1982. pp. 219–237. doi:10.1111/j.1365- 2818.1982.tb04625.x 4. Jeng T-W, Chiu W. Quantitative assessment of radiation damage in a thin protein crystal. Journal of Microscopy. 1984. pp. 35–44. doi:10.1111/j.1365-2818.1984.tb02544.x 5. Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods. 2013. pp. 584–590. doi:10.1038/nmeth.2472 6. Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, Veesler D, et al. Movies of Ice-Embedded Particles Enhance Resolution in Electron Cryo-Microscopy. Structure. 2012. pp.1823–1828. doi:10.1016/j.str.2012.08.026 7. De Rosier DJ, Klug A. Reconstruction of Three Dimensional Structures from Electron Micrographs. Nature. 1968. pp. 130–134. doi:10.1038/217130a0 8. Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157: 38–46. 9. Scheres SHW. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016;579: 125–157. 10. Punjani A, Fleet DJ. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J Struct Biol. 2021;213: 107702. 11. Zhao Y, Schmid MF, Frydman J, Chiu W. CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin. Nat Commun. 2021;12: 4754. 12. Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351: 969–972. 13. Rigort A, Bäuerlein FJB, Villa E, Eibauer M, Laugks T, Baumeister W, et al. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci U S A. 2012;109: 4449–4454. 14. Li Y, Huang W, Li Y, Chiu W, Cui Y. Opportunities for Cryogenic Electron Microscopy in Materials Science and Nanoscience. ACS Nano. 2020;14: 9263–9276. 15. Zhang Z, Cui Y, Vila R, Li Y, Zhang W, Zhou W, et al. Cryogenic Electron Microscopy for Energy Materials. Acc Chem Res. 2021;54: 3505–3517.
Biosketch

Wah Chiu is the Wallenberg-Bienenstock Professor in the Department of Bioengineering, Department of Microbiology and Immunology and the SLAC National Accelerator Laboratory at Stanford University. Formerly, he spent three decades at Baylor College of Medicine and is a founder of the W. M. Keck Center for Quantitative Biomedical Sciences with multiple academic institutions in the Greater Houston Area. His work has made multiple transformational contributions in developing single particle electron cryo-microscopy as a tool for the structural determination of molecular machines towards atomic resolution. He has collaborated broadly with researchers around the globe on cryo-EM of viruses, chaperonins, ion channels, RNA-protein complexes and RNAs. His recent research is focused on solving RNA-only cryo-Electron Microscopy structures relevant to infectious diseases and using cryo-Electron Tomography to study eukaryotic cell structures under different physiological conditions. He currently directs the Stanford-SLAC Cryo-EM Center https://cryoem.slac.stanford.edu/s2c2/, which is accessible to the global community for high resolution data collection and conducts training on cryo-EM methodology to new investigators.
Submit your questions for a panel discussion
